Multilayer Perceptron for squamous cell nuclei detection in Pap smear tests for cervical cancer diagnosis using local features
DOI:
https://doi.org/10.17488/RMIB.47.1.1537Palabras clave:
cervical cancer, multilayer perceptron , nuclei, squamous cellResumen
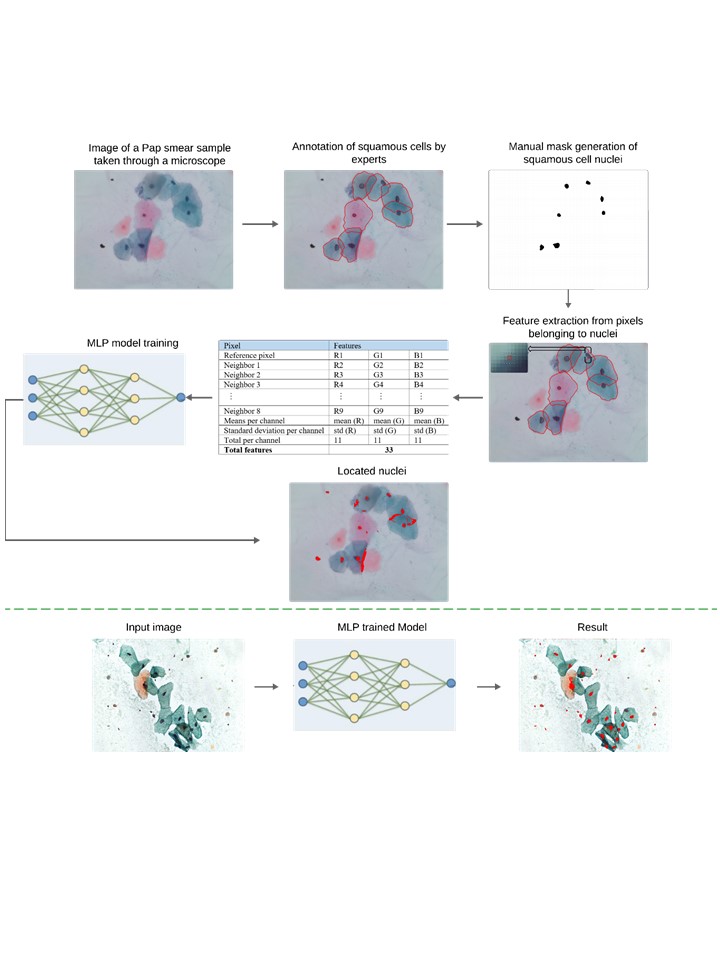
El cáncer cervicouterino es el cuarto tipo de cáncer más frecuentemente diagnosticado en mujeres a nivel mundial y representa una de las principales causas de muerte entre esta población. El diagnóstico se realiza manualmente a través del análisis de las células en las pruebas de Papanicolaou (Pap), lo que ocasiona retrasos en los resultados y una alta tasa de falsos positivos. Para abordar esta problemática, se propone un modelo de red neuronal artificial tipo Perceptrón Multicapa (MLP), que utiliza características locales para clasificar automáticamente los núcleos de células escamosas. El modelo fue entrenado con dos conjuntos de datos públicos: CRIC y SIPaKMeD. A diferencia de otros trabajos en el estado del arte, nuestro modelo aprende de la información de imágenes completas alcanzando una precisión de 0.9526 y 0.9397 en los conjuntos de datos CRIC y Sipakmed, respectivamente. Nuestra propuesta ofrece un enfoque sencillo mientras mantiene un rendimiento comparable al de los modelos complejos reportados en la literatura.
Descargas
Citas
073 Regional Committee for the Western Pacific, “Cervical cancer.” WHO Regional Office for the Western Pacific, p. Provisional agenda item 10, 2022.
72nd session Regional Committee for Europe, “Seventy-second Regional Committee for Europe: Tel Aviv, 12– 14 September 2022: roadmap to accelerate the elimination of cervical cancer as a public health problem in the WHO European Region 2022–2030.” World Health Organization. Regional Office for Europe, p. 8 p., 2022.
G. Bogani et al., “Duration of human papillomavirus persistence and its relationship with recurrent cervical dysplasia,” Eur. J. Cancer Prev., vol. 32, no. 6, pp. 525–532, 2023.
J. Lu, S. Han, J. Na, Y. Li, J. Wang, and X. Wang, “The correlation between multiple HPV infections and the occurrence, development, and prognosis of cervical cancer,” Front. Microbiol., vol. 14, p. 1220522, 2023.
L. A. Liang et al., “Cervical cancer screening: comparison of conventional Pap smear test, liquid-based cytology, and human papillomavirus testing as stand-alone or cotesting strategies,” Cancer Epidemiol. Biomarkers Prev., vol. 30, no. 3, pp. 474–484, 2021.
M. T. Corkum et al., “When pap testing fails to prevent cervix cancer: a qualitative study of the experience of screened women under 50 with advanced cervix cancer in Canada,” Cureus, vol. 11, no. 1, 2019.
P. Dilip Nandanwar, V. M. Wadhai, A. S. Chanchlani, and V. M. Thakare, “Analysis of Pixel Intensity Variation by Performing Morphological Operations for Image Segmentation On Cervical Cancer Pap Smear Image,” in 2021 International Conference on Computational Intelligence and Computing Applications (ICCICA), Nov. 2021, pp. 1–6. doi: https://doi.org/10.1109/ICCICA52458.2021.9697185
N. A. Alias, W. A. Mustafa, M. A. Jamlos, S. Ismail, H. Alquran, and M. N. K. H. Rohani, “Pap Smear Image Analysis Based on Nucleus Segmentation and Deep Learning–A Recent Review,” J. Adv. Res. Appl. Sci. Eng. Technol., vol. 29, no. 3, pp. 37–47, 2023.
S. Haridas and J. T, “Chaos and Exponential Scale based Butterfly Optimization Technique for Feature Extraction and Selection in Pap Smear Images,” in 2023 10th International Conference on Computing for Sustainable Global Development (INDIACom), Mar. 2023, pp. 740–744. Accessed: Feb. 04, 2025. [Online]. Available: https://ieeexplore.ieee.org/document/10112370
T. Chankong, N. Theera-Umpon, and S. Auephanwiriyakul, “Automatic cervical cell segmentation and classification in Pap smears,” Comput. Methods Programs Biomed., vol. 113, no. 2, pp. 539–556, 2014, doi: https://doi.org/10.1016/j.cmpb.2013.12.012
S. Dasgupta, “The Efficiency of Cervical Pap and Comparison of Conventional Pap Smear and Liquid-Based Cytology: A Review,” Cureus, vol. 15, no. 11, 2023.
M. Sangworasil et al., “Automated Screening of Cervical Cancer Cell Images,” in 2018 11th Biomedical Engineering International Conference (BMEiCON), Nov. 2018, pp. 1–4. doi: https://doi.org/10.1109/BMEiCON.2018.8609958
B. Z. Wubineh, A. Rusiecki, and K. Halawa, “Segmentation and Classification Techniques for Pap Smear Images in Detecting Cervical Cancer: A Systematic Review,” IEEE Access, vol. 12, pp. 118195–118213, 2024, DOI: https://doi.org/10.1109/ACCESS.2024.3447887
W. William, A. H. Basaza-Ejiri, J. Obungoloch, and A. Ware, “A Review of Applications of Image Analysis and Machine Learning Techniques in Automated Diagnosis and Classification of Cervical Cancer from Pap-smear Images,” in 2018 IST-Africa Week Conference (IST-Africa), May 2018, p. Page 1 of 11-Page 11 of 11. Accessed: Feb. 04, 2025. [Online]. Available: https://ieeexplore.ieee.org/document/8417373
N. Sompawong et al., “Automated Pap Smear Cervical Cancer Screening Using Deep Learning,” in 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jul. 2019, pp. 7044–7048. doi: https://doi.org/10.1109/EMBC.2019.8856369
J. S and D. F. X. Christopher, “Battle Royale Optimization Algorithm for Automatic Threshold Selection in Cervical Pap Smear Images,” in 2023 International Conference on IoT, Communication and Automation Technology (ICICAT), Jun. 2023, pp. 1–5. doi: https://doi.org/10.1109/ICICAT57735.2023.10263770
Z. Lu et al., “Evaluation of Three Algorithms for the Segmentation of Overlapping Cervical Cells,” IEEE J. Biomed. Health Inform., vol. 21, no. 2, pp. 441–450, Mar. 2017, doi: https://doi.org/10.1109/JBHI.2016.2519686
N. Youneszade, M. Marjani, and C. P. Pei, “Deep Learning in Cervical Cancer Diagnosis: Architecture, Opportunities, and Open Research Challenges,” IEEE Access, vol. 11, pp. 6133–6149, 2023, doi: https://doi.org/10.1109/ACCESS.2023.3235833
C. Suguna and S. P. Balamurugan, “An Efficient Water Strider Algorithm with Auto Encoder for Cervical Cancer Diagnosis using Pap Smear Images,” in 2022 4th International Conference on Smart Systems and Inventive Technology (ICSSIT), Jan. 2022, pp. 1463–1468. doi: https://doi.org/10.1109/ICSSIT53264.2022.9716358
I. S. Maesaroh, K. N. Syaja’Ah, Y. S. Perkasa, S. Hadianti, D. Riana, and R. R. Nurmalasari, “Cervical Cancer Classification From Pap Smear Using XCeption Model,” in 2024 10th International Conference on Wireless and Telematics (ICWT), Jul. 2024, pp. 1–5. doi: https://doi.org/10.1109/ICWT62080.2024.10674721
B. S. Chandana, C. Kommana, G. S. Madhav, P. Basa Pati, T. Singh, and A. K, “Explainable Screening and Classification of Cervical Cancer Cells with Enhanced ResNet-50 and LIME,” in 2024 3rd International Conference for Innovation in Technology (INOCON), Mar. 2024, pp. 1–7. doi: https://doi.org/10.1109/INOCON60754.2024.10512322
E. Zhang et al., “Cervical cell nuclei segmentation based on GC-UNet,” Heliyon, vol. 9, no. 7, 2023.
H. Kaur, R. Sharma, and L. Kaur, “Automated Cervical Cancer Image Analysis using Deep Learning Techniques from Pap-Smear Images: A Literature Review,” in 2021 9th International Conference on Reliability, Infocom Technologies and Optimization (Trends and Future Directions) (ICRITO), Sep. 2021, pp. 1–7. doi: https://doi.org/10.1109/ICRITO51393.2021.9596102
D. Jia, Z. Li, and C. Zhang, “A Parametric Optimization Oriented, AFSA Based Random Forest Algorithm: Application to the Detection of Cervical Epithelial Cells,” IEEE Access, vol. 8, pp. 64891–64905, 2020, doi: https://doi.org/10.1109/ACCESS.2020.2984657
M. T. Rezende et al., “Cric searchable image database as a public platform for conventional pap smear cytology data,” Sci. Data, vol. 8, no. 1, pp. 1–8, 2021.
M. E. Plissiti, P. Dimitrakopoulos, G. Sfikas, C. Nikou, O. Krikoni, and A. Charchanti, “SIPaKMeD: A New Dataset for Feature and Image Based Classification of Normal and Pathological Cervical Cells in Pap Smear Images,” in 2018 25th IEEE International Conference on Image Processing (ICIP), 2018, pp. 3144–3148. doi: https://doi.org/10.1109/ICIP.2018.8451588
Z. Meng, Z. Zhao, F. Su, and W. Wang, “Adaptive Elastic Loss Based on Progressive Inter-Class Association for Cervical Histology Image Segmentation,” in ICASSP 2020 - 2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), May 2020, pp. 976–980. doi: https://doi.org/10.1109/ICASSP40776.2020.9053232
D. Somasundaram, S. Gnanasaravanan, and N. Madian, “Automatic segmentation of nuclei from pap smear cell images: A step toward cervical cancer screening,” Int. J. Imaging Syst. Technol., vol. 30, no. 4, pp. 1209–1219, 2020, doi: https://doi.org/10.1002/ima.22444
D. N. Diniz et al., “A Deep Learning Ensemble Method to Assist Cytopathologists in Pap Test Image Classification,” J. Imaging, vol. 7, no. 7, Art. no. 7, Jul. 2021, doi: https://doi.org/10.3390/jimaging7070111
D. N. Diniz et al., “A Hierarchical Feature-Based Methodology to Perform Cervical Cancer Classification,” Appl. Sci., vol. 11, no. 9, Art. no. 9, Jan. 2021, doi: https://doi.org/10.3390/app11094091
“Introduction to Machine Learning,” MIT Press. Accessed: Sep. 25, 2024. [Online]. Available: https://mitpress.mit.edu/9780262043793/introduction-to-machine-learning/
C. M. Bishop, Pattern Recognition and Machine Learning. Springer, 2006.
D. Terra, A. Lisboa, M. Rezende, C. Carneiro, and A. Campos, “Shape-based Features Investigation for Preneoplastic Lesions on Cervical Cancer Diagnosis,” Feb. 2023. doi: https://doi.org/10.5220/0011900800003417
K. K. GV and G. M. Reddy, “Automatic Classification of Whole Slide Pap Smear Images Using CNN With PCA Based Feature Interpretation,” in 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Jun. 2019, pp. 1074–1079. doi: https://doi.org/10.1109/CVPRW.2019.00140
S. Targ, D. Almeida, and K. Lyman, “Resnet in Resnet: Generalizing Residual Architectures,” Mar. 25, 2016, arXiv: arXiv:1603.08029. doi: https://doi.org/10.48550/arXiv.1603.08029
National Cancer Institute (NCI), “The Bethesda System for Reporting Cervical Cytology: Definitions, Criteria, and Explanatory Notes”, 3rd ed. New York: Springer, 2015.
J. Long et al., "Fully Convolutional Networks for Semantic Segmentation," Proc. IEEE CVPR, pp. 3431–3440, 2015.
Descargas
Publicado
Cómo citar
Número
Sección
Licencia
Derechos de autor 2026 Revista Mexicana de Ingenieria Biomedica

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial 4.0.
Una vez que el artículo es aceptado para su publicación en la RMIB, se les solicitará al autor principal o de correspondencia que revisen y firman las cartas de cesión de derechos correspondientes para llevar a cabo la autorización para la publicación del artículo. En dicho documento se autoriza a la RMIB a publicar, en cualquier medio sin limitaciones y sin ningún costo. Los autores pueden reutilizar partes del artículo en otros documentos y reproducir parte o la totalidad para su uso personal siempre que se haga referencia bibliográfica al RMIB. No obstante, todo tipo de publicación fuera de las publicaciones académicas del autor correspondiente o para otro tipo de trabajos derivados y publicados necesitaran de un permiso escrito de la RMIB.











